企业性质生产商
入驻年限第9年
| 适用环境: | 实验室 |
PlantScreen高通量植物表型成像分析平台(传送带版)(二)
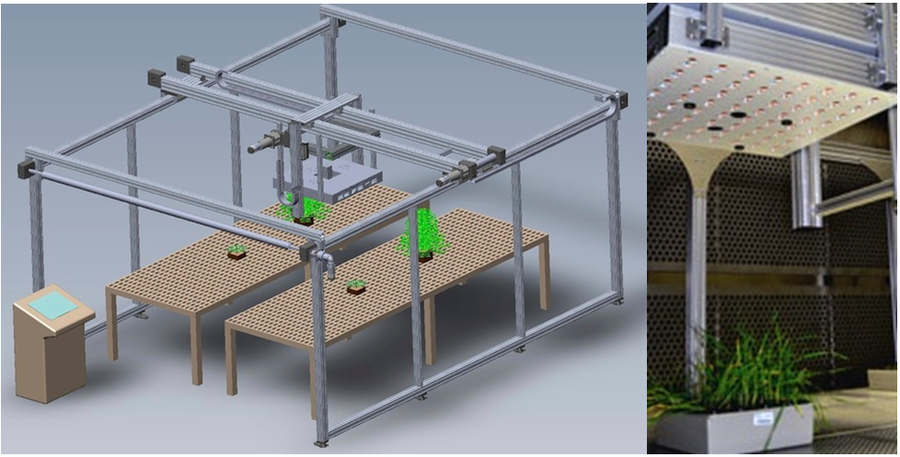
10.根系成像分析
·RhizoTron根窗技术,全自动成像分析,标配根窗44x29.5x5.8cm(高x宽x厚度)
·不仅可对根系成像分析,还可对地上苗(shoot)进行成像分析,苗高ZD50cm
·新一代CMOS传感器,分辨率12.3MP
·均一LED光源
·3层定位(顶部、中部、底部)根系浇灌系统(选配),3个水箱独立运行
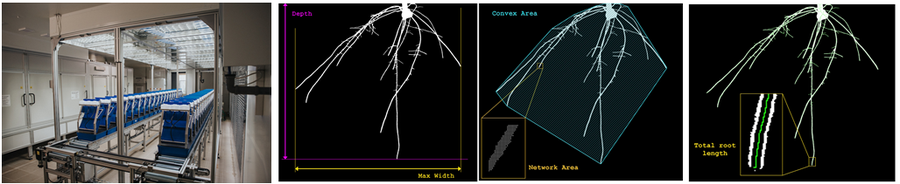
·测量参数包括:根深(或高度)、根冠宽度、高度与宽度比值、根冠面积、根冠紧实度、根系总长、轴对称性、根尖数、根节数等
11. 自动浇灌与称重单元
自动浇灌与称重单元
·测量参数:实际重量、浇水体积、ZZ重量、每个培养盆的相对重量
·操作指令:每个培养盆浇相同量的水(JD克数或者实际重量的百分比);保持相对重量;自定义每个培养盆的浇灌量模拟不同干旱或者内涝胁迫;称重前自动零校准,还可通过已知重量(如砝码)物品自动进行再校准
·每个培养盆的浇水量、日期、时间可分别程序控制记录以创建不同干旱胁迫梯度等,并且与整个系统的表型大数据无缝结合分析
·称重精度:大型植物±2g,小型植物±0.2g
·浇灌单元:流速3L/min,浇灌口高度可自动上下前后调整,保证ZJ浇灌位置
12.自动化植物传送系统
· 传送植物大小:根据客户需求,ZG可达200cm
传送植物大小:根据客户需求,ZG可达200cm
·传送带容纳量:50盆植物(1000株小型植物),可扩展100盆、200盆、400盆等更大容量 ;表型分析通量依不同的protocol而定,100分钟可以完成整个系统载荷植物样品的表型分析,可随机传送成像室进行成像分析、随机浇灌
·培养盆:防UV聚丙烯材料,标准5L(口径24cm)培养盆,可通过适配器应用3L培养盆,可360度旋转
·具备手动载样环(manual loading loop)以便在系统待机模式下手动载样分析实验、小组实验分析等
·具备激光植物高度测量监测系统和激光定位系统
·环形传送通道:具变速箱的三相异步马达,功率200-1000W,ZD负载500kg,速度150mm/s,传送带材料为防UV高耐用PVC
·移动控制系统:ZY处理单元CJ2M-CPU33;数字输入/输出ZD2560点;输入/输出单元ZD40;温度传感器Pt1000,Pt100,PTC;PLC通讯百兆以太网;OMRON MECHATROLINK-II ZD16轴精确定位
·RFID标签和QR植物辨识系统,自动读取每个样品托盘上的二维编码;辨识距离2-20cm;通讯RS485;可读取1维、2维和QR码;配备LED光源便于弱光下辨识
·环境监测传感器:温湿度传感器、PAR光合有效辐射传感器
·由主控制系统分别自动调控每一个样品托盘的测量时间、测量顺序、测量参数、浇灌时间和浇灌量,从测量单元到培养室的样品运转整个过程可实现完全自动控制,在无人值守情况下根据预设程序自行完成全部实验测量工作。
13.主控制表型大数据平台
·组成:控制调度服务器、客户端应用服务器、数据服务器、可编程序逻辑控制器及专业分析软件等,数据容量12TB
·自动控制与分析功能:具备用户定义、可编辑自动测量程序(protocols),根据用户设定程序自动完成全部实验。数据结果自动存储并分析,分析的数据结果可自动以动态曲线的形式显示。
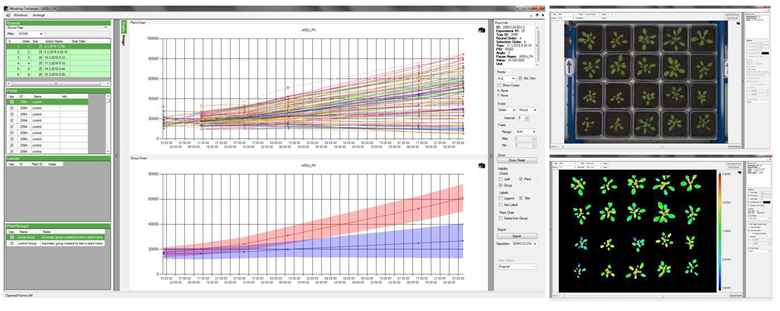
·MySQL数据库管理系统,可以处理拥有上千万条记录的大型数据库,支持多种存储引擎,相关数据自动存储于数据库中的不同表中
·植物编码注册功能:包括植物识别码、所在托盘的识别码等存储在数据库中,测量时自动提取自动读取条形码或RFID标签
·触摸屏操作界面,在线显示植物托盘数量、光线强度、分析测量状态及结果等,轻松通过软件完全控制所有的机械部件和成像工作站
·可用默认程序进行所有测量,也可通过开发工具创建自定义的工作过程,或者手动操作LED光源开启或关闭、RGB成像、叶绿素荧光成像、高光谱成像、红外热成像、3D激光扫描、称重及浇灌等
·叶片跟踪监测功能(leaf tracking)模块,可以持续跟踪监测叶片的生长、变化等等
·3D投射技术,可以通过高分辨率RGB镜头 或激光扫描构建3D模型,通过投射技术,将与其它传感器所得数据如叶绿素荧光、红外热成像温度数据、近红外数据、高光谱数据等投射在3D模型上一起进行对比分析等
·允许用户通过互联网远程访问,进行数据处理、下载及更改实验设计
·所测量的所有数据都是透明的、可以追溯的
·具备用户权限分级功能,防止其他人员误操作影响实验
·厂家远程故障诊断,软件终身免费升级

执行标准:
·CE认证标准
·CSN EN 60529 防护等级标准
·CSN 33 01 65 导体侧识别标准
·CSN 33 2000-3 基础特性标准
·CSN 33 2000-4-41ed.2 电击保护标准
·CSN 33 2000-4-43 电源过载保护标准
·CSN 33 2000-5-51ed.2 通用规则标准
·CSN 33 2000-5-523 容许电流标准
·CSN 33 2000-5-54ed.2 接地与保护导体标准
·CSN EN 55011 工业、科学与医学设备测量电磁干扰的范围与方法
·2006/42/EG 机械指令标准
·73/23/EEG 低电压指令标准
·2004/108/EG 电磁相容性指令标准
附:部分参考文献
1.M. Sorrentino, G. Colla, Y. Rouphaelouphael, K. Panzarová, M. Trtílek. 2020. Lettuce reaction reaction to drought stress: automated high-throughput phenotyping of plant growth and photosynthetic performance. ISHS Acta Horticulturae 1268.
2.Adhikari, P., Adhikari, T. B., Louws, F.F. J., & Panthee, D. R. 2020. Advances and Challenges in Bacterial Spot Resistance Breeding in Tomato (Solanum lycopersicum L.). International Journal of Molecular Sciences, 21(5), 1734.
3.Yang, W., Feng, H., Zhang, X., Zhang, J., Doonan, J. H., Et Al. 2020. Crop Phenomics and High-throughput Phenotyping: Past Decades, Current rent Challenges and Future Perspectives. Molecular Plant, 13(2), 187-214
4.Husičková, A., Humplík, J. F., Hýbl, M.,M., Spíchal, L., & Lazár, D. 2019. Analysis of Cold-Developed vs. Cold-Acclimated Leaves Reveals Various Strategies of Cold Acclimation of Field Pea Cultivars. Remote Sensing, 11(24), 2964
5.Singh, A.K., Yadav, B.S., Dhanapal, S., Berliner, M., Finkelshtein, A., Chamovitz, D.A. 2019. CSN Subunit of COP9 Signalosome Temporally Buffers Response to Heat in Arabidopsis. Biomolecules 2019, 9, 805.
6.Janečková, H., Husičková, A., Lazár, D., Ferretti, U., Pospíšil, P., & Špundová, M. 2019. Exogenous application of cytokinin during dark senescence eliminates the acceleration of photosystem II impairment caused by chlorophyll b deficiency in barley. Plant Physiology and Biochemistry, 136, 43–51
7.Marchetti, C. F., Ugena, L., Humplík, J. F., Polák, M., et al. 2019. A Novel Image-Based Screening Method to Study Water-Deficit Response and Recovery of Barley Populations Using Canopy Dynamics Phenotyping and Simple Metabolite Profiling. Frontiers in Plant Science, 10, 1252.
8.Rungrat T., Almonte A. A., Cheng R.,R., et al. 2019. A Genome-Wide Association Study of Non-Photochemical Quenching in response to local seasonal climates in Arabidopsis thaliana, Plant Direct, 3(5), e00138
9.Pavicic M, et al. 2019. High throughput invitro seed germination screen identifed new ABA responsive RING‑type ubiquitin E3 ligases inArabidopsis thaliana. Plant Cell, Tissue and Organ Culture 139: 563-575
10.Wen Z., et al. 2019. Chlorophyll fluorescence imaging for monitoring effects of Heterobasidion parviporum small secreted protein induced cell death and in planta defense gene expression. Fungal Genetics and Biology 126: 37-49
11.Gao G., Tester M. A., Julkowska M. 2019. The use of high throughput phenotyping for assessment of heat stress-induced changes in Arabidopsis. Biorvix, 838102.
12.Paul K., Sorrentino M., Lucini L., Rouphaelouphael Y. F., Cardarelli M., Bonini P., Begona M., Reyeynaud H.E., Canaguier R., Trtílek M., Panzarová K., Colla G. 2019. A Combined Phenotypic and Metabolomic Approach for Elucidating the Biostimulant Action of a Plant-derived Protein Hydrolysate on Tomato Grown un under Limited Water Availability. Frontiers in Plant Science, 10:493
13.Wang L., Poque S., Valkonen J. P. T. 2019. Phenotyping viral infection in sweetpotato using a high-throughput chlorophyll fluorescence and thermal imaging platform. Plant Methods, 15, 116
14.Paul K, Sorrentino M, Lucini L, Rouphaelouphael Y, Cardarelli M, Bonini P, Reynaud H,H, Canaguier R, Trtílek M, Panzarová K, Colla G. 2019. Understanding the Biostimulant Action of Vegetal-Derived Protein Hydrolysates by High-Throughput Plant Phenotyping and Metabolomics: A Case Study on Tomato. Frontiers in Plant Science, 10:47.
15.Gonzalez-Bayon, R., Shen, Y., Groszman, M., Zhu, A., Wang, A., et al. 2019. Senescence and defense pathways contribute to heterosis. Plant Physiology, 180, 240–252.
16.Julkowska, M. M., Saade, S., Agarwal Al, G., Gao, G., Pailles, Y., et al. 2019. MVApp–Multivaria analysis application for streamlined data analysis and curation. Plant Physiology, 180, 1261–1276.
17.Ganguly D. R., Stone B. A B., Eichten S. E., Pogson B. J. 2019. Excess light priming in Arabidopsis thaliana genotypes with altered DNA methylomes, G3: Genes, Genomes, Genetics, 9(11), 3611-3621
18.Ameztoy, K., Baslam, M., Sánchez-Lópeópez, Á. M., Muñoz, F. J., et al. 2019. Plant responses to fungal volatiles involve global post-translational thiol redox proteome changes that affect photosynthesis. Plant, Cell & Environment, 42(9), 2627-2644.
19.Adhikari N. D., Simko I., Mou B. 2019. Phenomic and Physiological Analysis of Salinity Effects on Lettuce. Sensors 19, 4814.
20.Ugena L, Hýlová A, Podlešáková K,K, Humplík J.F., Doležal K, Diego N, Spíchal L. 2018. Characterization of Biostimulant Mode of Action Using Novel Multi-Trait High-Throughput Screening of of Arabidopsis Germination and Rosette Growth. Frontiers in Plant Science, 9:1327.
21.Lyu, J. I., Kim, J. H., Chu, H., Taylor, M.M. A., Jung, S., et al. 2018. Natural allelic variation of GVS1 confers diversity in the regulation of leaf senescence in Arabidopsis. New Phytologist, 221(4), 2320-2334
22.Ganguly D. R., Crisp P. A., Eichten S. R., et al. 2018. Maintenance of pre-existing DNA methylation states through recurring excess-light stress. Plant Cell and Environment. 41(7), 1657-1672.
23.Rouphael Y., Spíchal L., Panzarová K.,K., et al. 2018. High-throughput Plant Phenotypin ping for Developing Novel Biostimulants: From Lab to Field or FroFrom Field to Lab? Front. Plant Sci., 9:1197.
24.Coe R. A., Chatterjee J., Acebron K., et al. 2018. High-throughput chlorophyll fluorescence screening of Setaria viridis for mutants with altered CO2 compensation points. Functional Plant Biology. 45(10), 1017-1025
25.Fichman Y., Koncz Z., Reznik N., et al. 2018. SELENOPROTEIN O is a chloroplast protein involved in ROS scavenging and its absence increases dehydration tolerance in Arabidopsis thaliana. Plant Science. 41(7), 1657-1672
26.Sytar O., Zivcak M., Olsovska K., Brestic M. 2018. Perspectives in High-Throughput Phenotyping of Qualitative Traits at the Whole-Plant Level. In: Sengar R., Singh A. eds Eco-friendly Agro-biological Techniques for Enhancing Crop Productivity. Springer, Singapore, 213-243.
27.De Diego N., Fürst T., Humplík J. F., et al. 2017. An Automated Method for High-Throughput Screening of Arabidopsis Rosette Growth in Multi-Well Plates and Its Validation in Stress Conditions. Frontiers in Plant Science. 8.
28.Lobos G. A., Camargo A. V., del Pozo A., et al. 2017. Editorial: Plant Phenotyping and Phenomics for Plant Breeding. Front. Plant Sci. 8.
29.Pavicic M., Mouhu K., Wang F., et al. 2017. Genomic and Phenomic Screens for Flower Related RING Type Ubiquitin E3 Ligases in Arabidopsis. Frontiers in Plant Scienc. Volume 8.
30.Rungrat T., Awlia M., Brown M. et al. 2017. Monitoring Photosynthesis by In Vivo Chlorophyll Fluorescence: Application to High-Throughput Plant Phenotyping. The Arabidopsis Book 14: e0185. 2016
31.Simko I., Hayes R. J. and Furbank R. T. 2017. Non-destructive Phenotyping of Lettuce Plants in Early Stages of Development with Optical Sensors. Frontiers in Plant Science. 2016;7:1985.
32.Sytar O., Brestic M., Zivcak M., et al. 2017. Applying hyperspectral imaging to explore natural plant diversity towards improving salt stress tolerance. In Science of The Total Environment, 578, 90-99.
33.Sytar O., Brücková K., Kovár M., et al. 2017. Nondestructive detection and biochemical quantification of buckwheat leaves using visible VIS and near-infrared NIR hyperspectral reflectanceimaging. Journal of Central European Agriculture. 184, 864-878
34.Tschiersch H., Junker A., Meyer R. C., & Altmann, T. 2017. Establishment of integrated protocols for automated high throughput kinetic chlorophyll fluorescence analyses. Plant Methods, 13, 54.
35.Weber J., Kunz, C., Peteinatos, G., et al. 2017. Utilization of Chlorophyll Fluorescence Imaging Technology to Detect Plant Injury by Herbicides in Sugar Beet and Soybean. Weed Technology, 1-13.
36.Awlia M., Nigro A., Fajkus J., Schmöckel S.M., Negrão S., Santelia D., Trtílek M., Tester M., Julkowska M.M. and Panzarová K. 2016: High-throughput non-destructive phenotyping of traits contributing to salinity tolerance in Arabidopsis thaliana. Submitted Frontiers in Plant Sciences.
37.Bell J. and Dee M. H. 2016. The subset-matched Jaccard index for evaluation of Segmentation for Plant Images. Front Plant Sci. 2016; 7: 1985.
38.Bell J. and Dee M. H. 2016. Watching plants grow – a position paper on computer vision and Arabidopsis thaliana. IET Computer Vision. Volume 11, Issue 2, March 2017, p. 113 – 121.
39.Bush M.S., Pierrat O, Nibau C, et al.2016. eIF4A RNA Helicase Associates with Cyclin-Dependent Protein Kinase A in Proliferating Cells and is Modulated by Phosphorylation. Plant Physiol. 2016 Jul 7,
40.Cruz J. A., Savage L. J., Zegarac R., et al. 2016. Dynamic Environmental Photosynthetic Imaging Reveals Emergent Phenotypes. Cell Systems, Volume 2, Issue 6, 2016, Pages 365-377.
41.Sytar O., Brestic M., Zivcak M . 2016. Noninvasive Methods to Support Metabolomic Studies Targeted at Plant Phenolics for Food and Medicinal Use. Plant Omics: Trends and Applications.
42.Humplik J.F., Lazar D., Husickova A. and Spichal L. 2015: Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses – a review. Plant Methods 11:29.
43.Humplik J.F., Lazar D., Fürst, T., Husickova A., Hybl, M. and Spichal L. 2015: Automated integrative high-throughput phenotyping of plant shoots: a case study of the cold-tolerance of pea Pisum sativum L.. Plant Methods 19;11:20.
44.Brown T.B., Cheng R., Sirault R.R., Rungrat T., Murray K.D., Trtilek M., Furbank R.T., Badger M., Pogson B.J., and Borevitz J.O. 2014: TraitCapture: genomic and environment modelling of plant phenomic data. Current Opinion in Plant Biology 18: pp. 73-79.
45.Mariam Awlia, et.al, 2016, High-Throughput Non-destructive Phenotyping of Traits that Contribute to Salinity Tolerance in Arabidopsis thaliana, Frontiers in Plant Science, DOI: 10.3389/fpls.2016.01414
46.Ivan Simko, et.al, 2016, Phenomic approaches and tools for phytopathologists, Phytopathology, DOI: 10.1094/PHYTO-02-16-0082-RVW
47.Tepsuda Rungrat, et.al, 2016, Using Phenomic Analysis of Photosynthetic Function for Abiotic Stress Response Gene Discovery, The Arabidopsis Book 14: e0185, The American Society of Plant Biologists, DOI: http://dx.doi.org/10.1199/tab.0185
48.Jorge Marques da Silva, 2016, Monitoring Photosynthesis by In Vivo Chlorophyll Fluorescence: Application to High-Throughput Plant Phenotyping, Applied Photosynthesis - New Progress, Edition 1, Chapter 1, pp:3-22, DOI: http://dx.doi.org/10.5772/62391
49.Maxwell S. Bush, et.al, 2016, eIF4A RNA Helicase Associates with Cyclin-Dependent Protein Kinase A in Proliferating Cells and is Modulated by Phosphorylation. Plant Physiol., DOI: 10.1104/pp.16.00435
50.Ángela María Sánchez-López, et.al, 2016, Volatile compounds emitted by diverse phytopathogenic microorganisms promote plant growth and flowering through cytokinin action, Plant, Cell and Environment, DOI: 10.1111/pce.12759
51.Jan Humplík, et.al, 2015, Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses – a review, Plant Methods, 11: 29
52.Jan Humplík, et.al, 2015, Automated integrative high-throughput phenotyping of plant shoots: a case study of the cold-tolerance of pea Pisum sativum L., Plant Methods, 11: 20
53.Pip Wilson, et.al, 2015, Genomic Diversity and Climate Adaptation in Brachypodium, Chapter Genetics and Genomics of Brachypodium, Volume 18 of the series Plant Genetics and Genomics: Crops and Models, pp:107-127
54.Tim Brown, et.al, 2014, TraitCapture: genomic and environment modelling of plant phenomic data, Current Opinion in Plant Biology, 18: 73-79
55. Jan Humplík, et.al, 2014, High-throughput plant phenntyping facility in Palacky University in Olomouc, International Symposium on Auxins and Cytokinins in Plant Development
附:其它表型分析平台:
1、FKM多光谱荧光动态显微成像系统
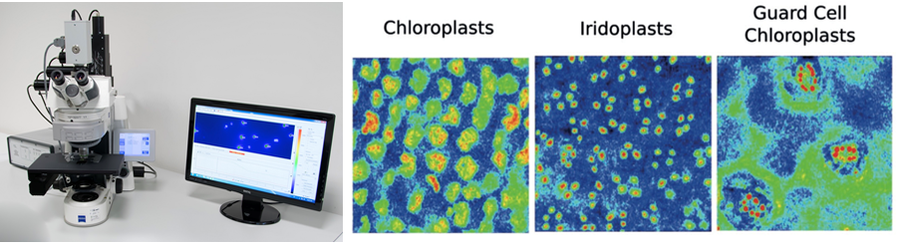
右图引自《Nature Plants》2016, Photonic multilayer structure of Begonia chloroplasts enhances photosynthetic efficiency by Heather M. Whitney等
2、PlantScreen-R移动式表型分析平台(下左图):用于大田植物叶绿素荧光成像分析、RGB成像分析、红外热成像分析、3D激光扫描测量分析等

3、PlantScreen台式及移动式植物表型分析平台(参见上右图)
1)3D RGB彩色成像分析
2)FluorCam叶绿素荧光成像分析
3)FluorCam多光谱荧光成像分析
4)高光谱成像分析
5)红外热成像分析
6)PAR吸收/NDVI成像分析
7)近红外3D成像分析
4、PlantScreen样带式表型分析平台

5、PlantScreen 植物表型三维自动扫描成像分析平台